iGEM REPORT: Kinetic Modeling of a Novel Protein Detection System Using Proximity-Dependent Ligation and Nucleic Acid Sequence-Based Amplification
Note: This iGEM Report was submitted to the PLOS iGEM Realtime Peer Review Jamboree, and has not undergone formal peer review by any of the PLOS journals. We welcome your comments on this work.
Kinetic Modeling of a Novel Protein Detection System Using Proximity-Dependent Ligation and Nucleic Acid Sequence-Based Amplification
Nick Emery (1) *, Aaron Renberg (1), Cristina Castillo (2), Akira Nishii (1,3), Alex Hadd (3), Alex Girgis (3), Zach Bernstein (1), Daniela Lewin Mejia (1), Caleb Kaczmarek (1), Robbin Pearce (3), Vijay Vobbilisetty (3), Irina Kopyeva (3), Rachel Sun (1)
- University of Michigan College of Literature, Science, and the Arts
- University of Michigan Department of Molecular Integrative Physiology
- University of Michigan College of Engineering
*Corresponding Author: Nick Emery (emernic@umich.edu)
Author Contributions
Conceptualization: NE CC
Methodology: AN NE CC
Software: NE AN
Formal Analysis: NE AN
Investigation: AR NE CC AH AG ZB DL CK RP VV IK AN RS
Resources: RS
Writing-Original Draft: AR AN ER CC
Writing- Review and Editing: NE AR VV AG AN
Visualization: VV
Supervision: AR NE CC RS
Project Administration: RS CC DL AH
Funding Acquisition: VV AG AN RS AR CC NE
Abstract
Using aptamer-based proximity-dependent ligation (PDL), nucleic acid sequence based amplification (NASBA), and cell-free expression, we designed a novel paper-based diagnostic device that produces a colorimetric output in the presence of a target protein biomarker. This system, which could be performed outside the lab with minimal training, could provide an inexpensive alternative to enzyme-linked immunosorbent assays (ELISA). The reaction scheme begins with two single-stranded oligonucleotide probes which selectively bind to independent sites on a protein biomarker via aptamer domains. The selective binding of the oligonucleotide probes to the target protein promotes the ligation of the two probes into one continuous ssDNA strand by T4 DNA ligase. The oligonucleotide formed by the ligation of the probes is amplified exponentially via a modified NASBA reaction to increase the signal, enabling lower detection limits for protein biomarkers. After amplification, the alpha fragment of LacZ is expressed through a cell-free expression reaction. The alpha fragment can then undergo alpha complementation with the LacZ omega fragment present in the reaction mixture, forming functional LacZ capable of cleaving X-gal, forming a colorimetric output. The system is designed to operate as a one-pot reaction under isothermal conditions and would be freeze-dried onto paper for use as a point-of-care device. By using aptamer-based PDL as the foundation for the detection scheme, this system could potentially attain the extremely low detection limits characteristic of PCR-coupled PDL in an inexpensive, easily portable system. To assess the theoretical viability of this scheme, we created a kinetic model of the proximity-dependent ligation reaction, using thrombin as a model protein biomarker. The modeling results indicate that such a system could give a linear response with respect to initial biomarker concentration over at least five orders of magnitude. Preliminary in vitro testing of the proximity-dependent ligation reaction using thrombin was also performed but was inconclusive.
Financial Disclosure
The Michigan Synthetic Biology Team collectively received funds from: University of Michigan Central Student Government; University of Michigan Program in Biology; University of Michigan College of Engineering; Cayman Chemical; University of Michigan College of LS&A OptiMize Department; University of Michigan Women in Science & Engineering. This work was in part funded using the science crowdfunding platform experiment.com. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Data Availability
All data are fully available without restriction.
Introduction
Accurate, rapid, and affordable disease diagnosis in regions with little health care infrastructure remains a major challenge in the world. We set out to solve this major global issue using cell-free synthetic biology as a platform. It has been previously demonstrated that RNA biomarkers can be detected in a cell-free expression reaction using a toehold riboswitch, and that such detection systems can be freeze-dried onto paper for later use [1,2]. A major obstacle to the efficacy of these systems for real-world diagnostics is a lack of signal amplification. While PCR or RT-PCR can amplify nucleic acid biomarkers in a laboratory setting, these methods are impractical in the poor infrastructure areas where this paper-based technology would be most appealing. To combat this, we turned to a modified form of proximity-dependent ligation, followed by NASBA. Proximity-dependent ligation involves two single stranded DNA probes and a ssDNA “bridge” oligo. The two probes are each designed with a protein-specific aptamer on one end, and half of a reporter gene sequence on the other. The bridge segment is complementary to the non-aptamer ends of the two probes. As seen in Figure 1, when the two probes come in contact with the target protein, their aptamer domains bind to it. This increases the effective concentrations of their non-aptamer ends, allowing the bridge segment to anneal to the two free ends of the probes, connecting them. This complex is ligated by T4 DNA Ligase, allowing the reporter gene to be transcribed by T7 RNA Polymerase. The key feature of this modified PDL system is that the whole reporter gene is only transcribed in the presence of the target protein [3]. In order to increase signal amplification, the ligated ssDNA template is amplified by NASBA, as seen in Figure 2, which allows PCR-like amplification at a single temperature in a one-pot reaction [4]. This whole system could be freeze-dried onto paper and used as a diagnostic device for the detection of protein biomarkers without the need for advanced laboratory infrastructure.
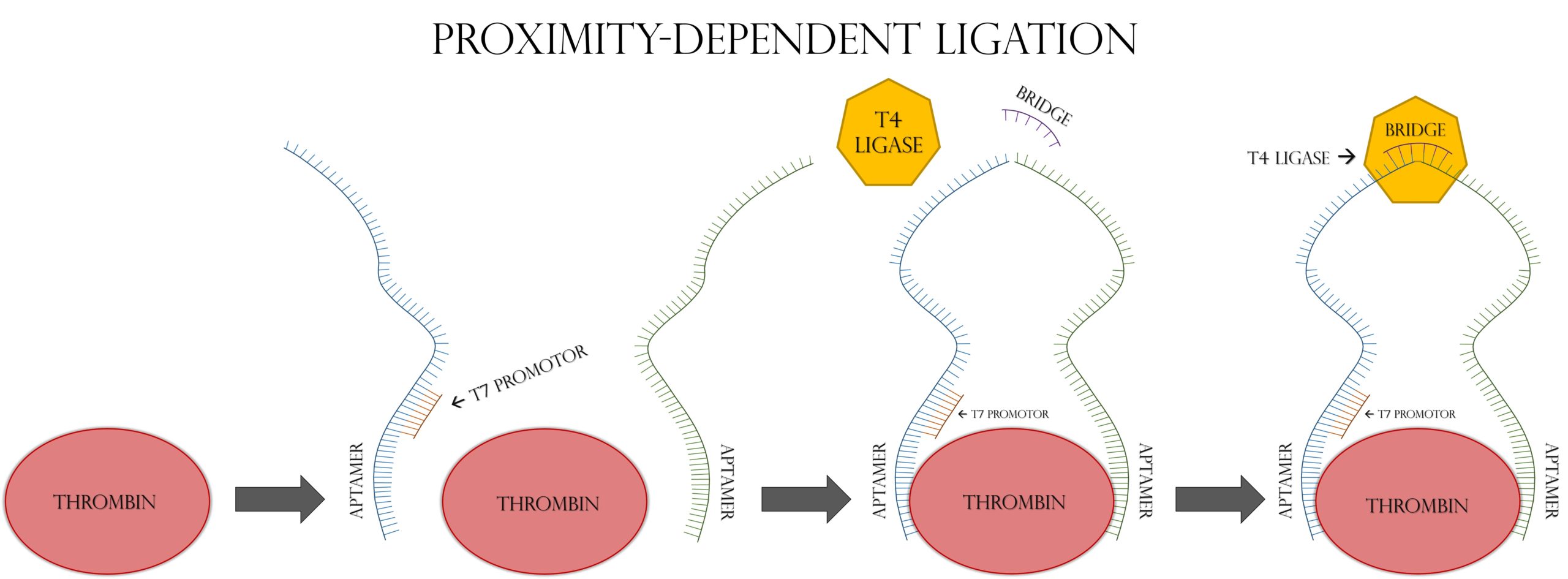
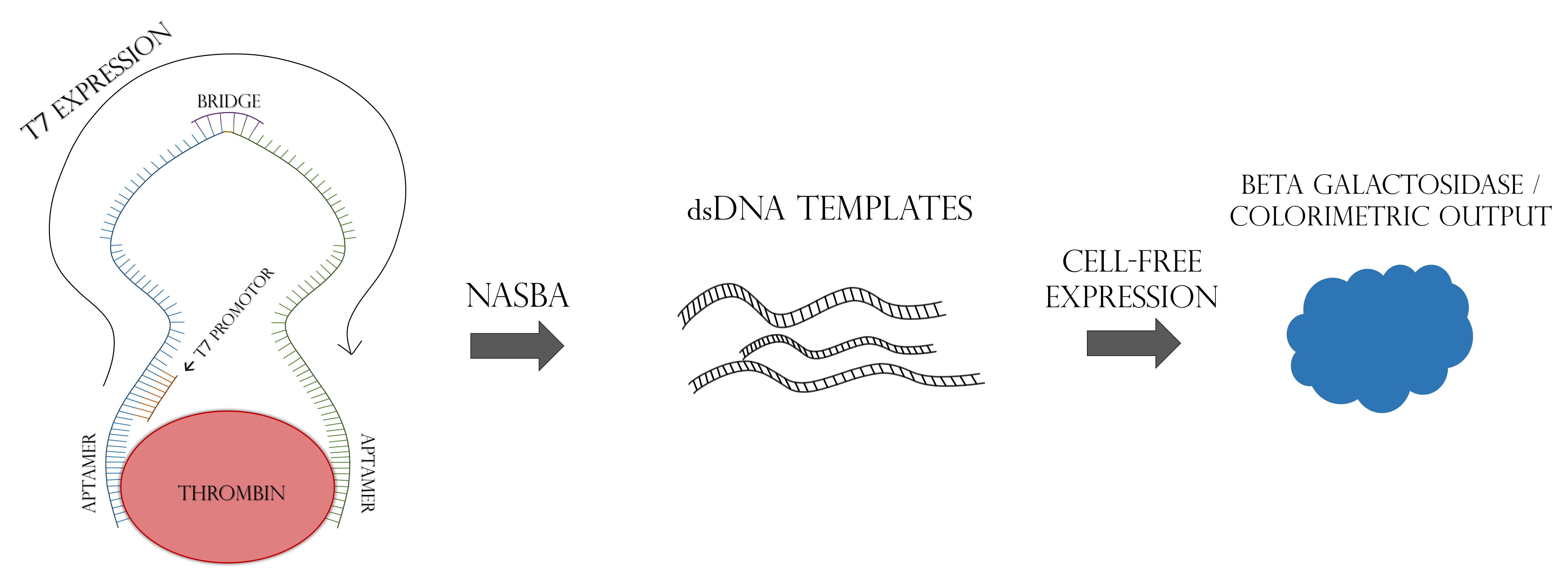
Methods
Materials
T4 DNA Ligase and restriction enzymes used for cloning were purchased from New England Biolabs. RNAse inhibitor was purchased from Roche and Thrombin was purchased from Haematologic technologies Inc. All oligonucleotides were purchased from Integrated DNA Technologies Inc.
Plasmid Cloning
The lacZ gene from iGEM biobrick BBa_K564012 was subcloned into the PET28a. Overhang Chain Polymerase reaction was performed on the lacZ sequence adding EcoRI and XbaI upstream and HindIII and PstI downstream. The PCR output was then digested with EcoRI and HindIII and ligated into PET28a. This plasmid was used to create a delta M-15 mutant plasmid, which has amino acid residues 11-41 deleted, leaving only the lacZ omega fragment. This deletion was performed using the Q5 site direct mutagenesis kit from NEB.
Cell line
Sigma DH5alpha chemically competent cells were used for cloning purposes.
Mathematical Modeling
A mathematical model of the proximity-dependent ligation reaction was created using SimBiology version 5.4, an add-on for MatLab version R2016a. All binding events were modeled as reversible reactions with Kon rate constants set to 1010 M-1s-1 to approximate the diffusion limit in water. Koff rate constants for nucleic acid hybridizations were based on binding energies determined using the Mfold web server under the buffer conditions used in the proximity-dependent ligation reaction. Koff rate constants for aptamer-protein interactions were determined using Kd values reported for these aptamers in the literature [5,6]. T4 DNA Ligase kinetic parameters were obtained from the literature [7,8]. A complete copy of the kinetic model which includes all parameter values can be found here. Key assumptions of the model include: negligible protein degradation, diffusion-limited binding rates, and negligible off-target interactions between aptamers and other reaction components.
Results
A kinetic model involving 20 reaction species and 27 possible reactions was used to simulate the proximity-dependent ligation step of our amplification scheme and is illustrated in Figure 3.
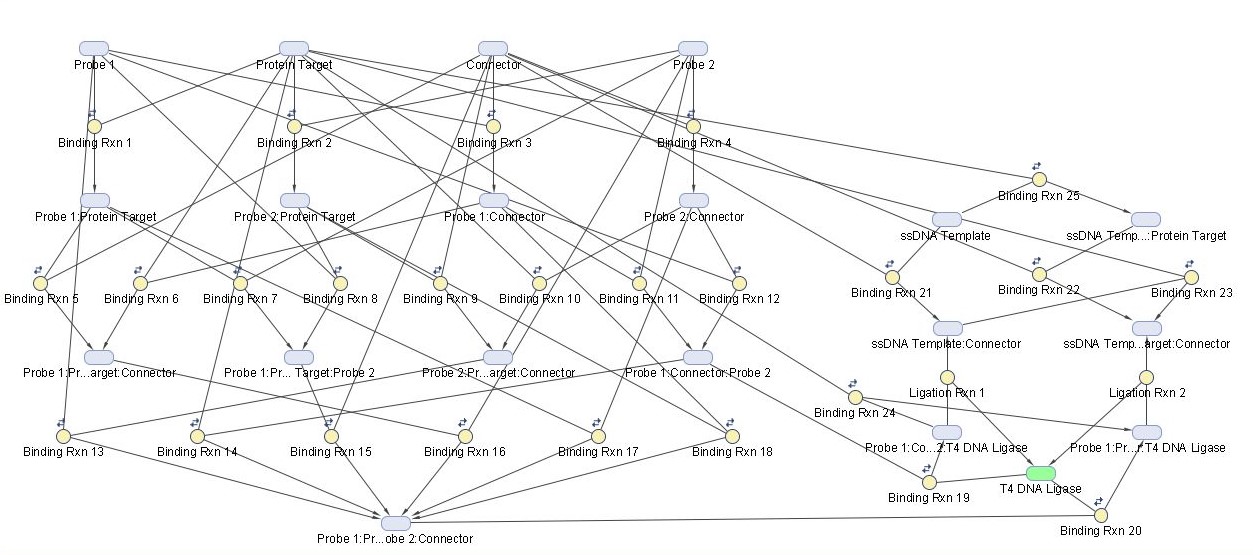
Kinetic parameters and underlying assumptions of the model were chosen as described in the methods section. Probe 1 and probe 2 represent the two aptamer-containing ssDNA probes which bind to the protein target. The connector species represents the ssDNA bridge oligonucleotide which weakly binds the non-aptamer ends of the two probes together for ligation by T4 DNA Ligase. Human Alpha Thrombin was chosen as a model protein target for this system due to the existence of well characterized aptamers for thrombin, which have been used for proximity-dependent ligation reaction in existing literature.
The reaction was simulated using 5 different initial concentrations of thrombin as well as a negative control. Initial concentrations of probe 1, probe 2, and connector were all set to 100 pM. A plot of the concentration of total ssDNA template, which is the product of ligation by T4 DNA Ligase, was generated in Figure 4.
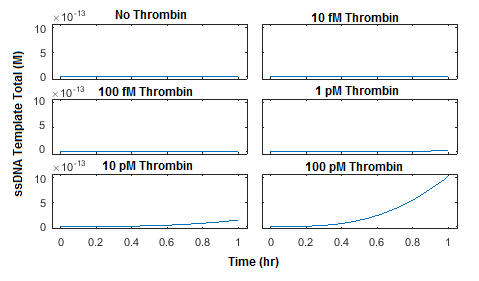
This simulation was then repeated using initial concentrations of probe 1, probe 2, and connector at 1 nM each (Figure 5) and 10 nM each (Figure 6). The plots show that increasing the concentration of probe 1, probe 2, and connector from 100 pM to 1 nM can increase the production of ligated ssDNA template by approximately two orders of magnitude. Increasing the concentrations of probe 1, probe 2, and connector further to 10 nM yields only a two-fold increase in ssDNA template produced and noticeably increases background ligation when thrombin is not present.
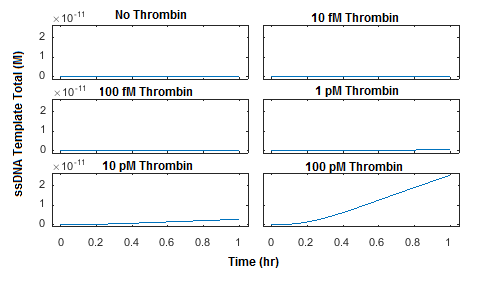
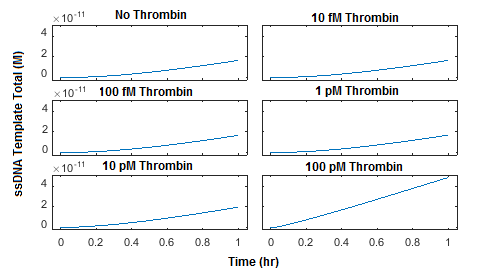
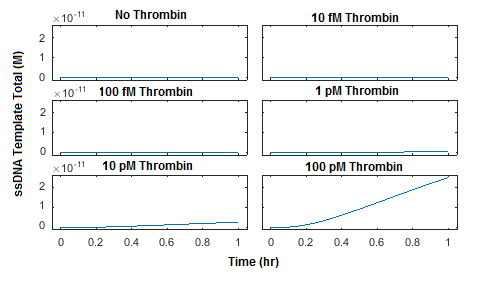
The model was also used to assess whether target “recycling” contributes significantly to the ligation reaction. Target recycling in this case refers to the possibility of a single molecule of protein target catalyzing multiple ligation reactions. In our model, this is represented by a reversible reaction in which a molecule of protein target comes unbound from the ligated ssDNA template, allowing it to be bound by another pair of ssDNA probes. The model used in Figures 3-6 allows for target recycling; therefore, to quantitatively assess the effect of recycling on the reaction rate, a new model was generated which does not allow for the protein target to come unbound from the ligated ssDNA template (Figure 7). The parameters used in this reaction were otherwise identical to those used in Figure 4. Interestingly, these results are nearly identical to those observed in Figure 4, suggesting that the effect of target recycling is essentially negligible. This observation is likely generalizable to all proximity-dependent ligation reactions with aptamer affinities that are similar or stronger than the thrombin aptamers used. This suggests that the maximum concentration of ligated ssDNA template produced by such a system cannot greatly exceed the initial concentration of protein target.
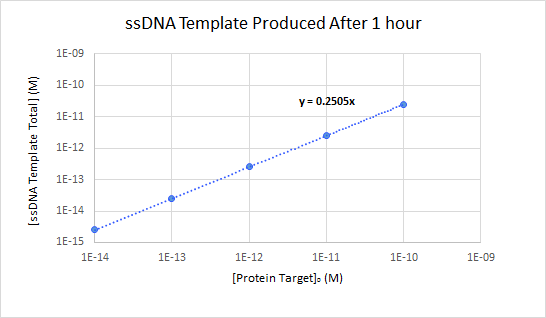
In order to assess the linearity of the relationship between initial protein target concentration and final ssDNA template concentration, a more detailed analysis was performed on the simulation shown in Figure 4. The final concentration of ssDNA template produced was plotted with respect to the initial concentration of thrombin (Figure 8). The run in which no thrombin was added was used for background subtraction. The results show a highly linear response for all concentrations of thrombin tested, with final ssDNA concentrations which were 25.05% of the initial thrombin concentrations.
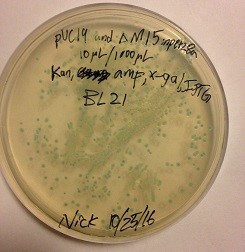
In addition to kinetic modeling, we also attempted to perform the proximity-dependent ligation reaction in-vitro using SYBR Gold agarose gels to detect the ligation product. The results were inconclusive (data not shown). This may have been due to unexpected behavior of the system at the high concentrations required for gel visualization, or a variety of factors including low specific activity of the protein target or an inadequate proportion of full-length probes due to limitations in oligonucleotide synthesis. A pET28a protein expression plasmid coding for the lacZ omega fragment was produced by subcloning the coding sequence of lacZ into pET28a and then performing a deletion mutagenesis to remove residues 11-41. This construct was shown to be capable of producing the LacZ omega fragment in vivo by co-transforming it with a separate plasmid expressing the LacZ alpha fragment (Figure 9). This cotransformation was done in BL21(DE3) cells which do not carry a chromosomal copy of either fragment of the lacZ gene. Transformation of either plasmid alone did not yield the characteristic blue color seen in the co-transformation (data not shown).
Discussion/Conclusions
The results generated through kinetic modeling are preliminary but encouraging. The model predicts final ssDNA template concentrations of up to 25% of the initial protein target concentration, and the relationship between ssDNA template produced and protein target added is highly linear over at least five orders of magnitude. This demonstrates that PLA is able to efficiently convert a protein signal, the concentration of thrombin, into a nucleic acid signal, the production of ssDNA template. This constitutes a key step in our reaction scheme because it allows us to take advantage of NASBA’s high signal amplification despite the fact that NASBA is designed to amplify nucleic acid targets rather than proteins. We found that using ~1 nM of probe 1, probe 2, and connector gave a strong and highly linear level of ligation in the presence of thrombin while maintaining a low background level of ligation in the absence of thrombin. It was also found that target recycling probably contributes very little to the overall reaction rate; therefore, a key limitation of this system would be that PLA will always produce less ligated ssDNA template than the initial concentration of protein target.
Future work would involve extending the model to encompass downstream reaction steps such as NASBA and cell-free expression. The model could also be generalized by testing a wide range of aptamer binding affinities for a variety of targets, and by altering the connector and probe sequences to change nucleic acid hybridization energies. We would also like to demonstrate this reaction scheme in solution and as a freeze-dried test strip for the detection of a clinically relevant protein biomarker. Successful demonstration of this reaction on paper could allow for the creation of a broad variety of paper-based assays for protein biomarkers, creating far-reaching positive impacts on diagnostic testing including lowering cost of production and consumption, minimizing the time it takes for a complete diagnostic test, and increasing diagnostic accuracy. This would be a major improvement for healthcare systems around the globe.
Acknowledgements
Special thanks to Marc Ammerlaan, Pearl Tu, OptiMize, Dennis Drobeck, iGEM, Caymen Chemical, and the University of Michigan.
Response to Reviewers
A transcript of the reviewer comments and author responses from the Live Peer Review Jamboree can be found here: Michigan Response to Reviewers
References
- Green ACAA, Silver PA, Collins JJ, Yin P. Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell. 2014;159(4):925–39.
- Pardee K, Green AA, Ferrante T, Cameron DE, Daleykeyser A, Yin P, et al. Paper-Based Synthetic Gene Networks. Cell. 2014;159(4):940–54.
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nature Biotechnology. 2002;20(5):473–7.
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991Jul;350(6313):91–2.
- Macaya RF, Waldron JA, Beutel BA, Gao H, Joesten ME, Yang M, et al. Structural and Functional Characterization of Potent Antithrombotic Oligonucleotides Possessing Both Quadruplex and Duplex Motifs. Biochemistry. 1995;34(13):4478–92.
- Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. Journal of Molecular Biology. 1997;272(5):688–98.
- Lohman GJS, Chen L, Evans TC. Kinetic Characterization of Single Strand Break Ligation in Duplex DNA by T4 DNA Ligase. Journal of Biological Chemistry. 2011;286(51):44187–96.
- Promega Enzyme Resource Guide. Promega; https://www.promega.com/-/media/files/resources/product-guides/cloning-enzymes/ligases.pdf?la=en
Supporting Information
Supporting information for this work can be found here: Michigan SI